Lightsheet - MesoSPIMs (Irchel)

The mesoSPIM (mesoscale selective plane illumination microscopy) is the ideal instrument to quickly bridge scales from the µm- to the cm-level, which enables it to serve as an excellent tool for detailed three-dimensional anatomical investigations in many biological and medical fields (Voigt et al., 2019, Vladimirov et al, 2024).
Main features of both setups (Benchtop and V6):
- Compatibility with all clearing techniques: Clarity, iDISCO, CUBIC, PEGASOS, ECI, etc.
- Large field-of-view, ranging from 1.5 to 15 mm using a set of Mitutoyo Plan Apo objectives.
- Large-sensor low-noise sCMOS cameras
- Dual-sided excitation. Optimized for fast screening of whole mouse brain samples, yielding relatively small datasets (10-14 GB/brain/color) near-isotropic sampling (2-4 µm in X,Y & Z) due to an axially swept lightsheet which leads to uniform z-resolution across the entire field-of-view.
- Modular sample holders and quick sample exchange.
- Large travel range (45 x 45 x 100 mm) to allow imaging of very large samples.
Further information on the UZH mesoSPIM open-source project: MesoSPIM.org
Location
University Zurich, Irchel Campus, Room Y44-J-33.
Training Request
Pleaseregister to apply for an introduction to the microscopes.
Introduction session duration: 2 hours
If you have used mesoSPIM V6 before and would like to use the Benchtop, please request a training session as well, the systems have some differences in the startup sequence and interface.
Technical Specifications
Excitation path
Both systems have excitation based on axially scanned light-sheet microscopy (ASLM). The waist of the light-sheet is translated through the sample in synchrony with the rolling shutter of the camera, which enables high axial resolution across very large fields of view.
Light Sources and Lasers
mesoSPIM V6: iChrome MLE
Benchtop: Oxxius L4Cc
Laser lines available on both systems
- 405nm (100mW)
- 488nm (100mW)
- 561nm (100mW)
- 640nm (100mW)
Detection path
Both systems have long-working distance air plan apochromat objectives:
- Mitutoyo M Plan Apo 2x/0.055
- Mitutoyo M Plan Apo 5x/0.14
- Mitutoyo M Plan Apo 7.5x/0.21
- Mitutoyo M Plan Apo 10x/0.28
- Mitutoyo M Plan Apo 20x/0.28(t3.5) (corrected for 3.5 mm thick glass)
The microscopes offers lateral (XY) resolution of 1.5-2.6 µm (depending on the objective) and axial (Z) resolution of 3.5 µm for all objectives, across a large field of view thanks to new-generation sCMOS cameras.
Emission (detection) filters
- Quadband filter 405/488/561/640 (both systems)
- 520/35 (mesoSPIM V6)
- 590/36 (mesoSPIM V6)
- 488 long-pass, AHF #F76-488 (Benchtop)
If you need other filters, please talk to us.
Cameras
| System | mesoSPIM V6 | Benchtop |
| Camera | Hamamatsu Orca Lightning | Photometrics Iris 15 |
| image size | 4608 x 2592 px (12 MP) | 5056 x 2960 px (15 MP) |
| pixel size | 5.5 µm | 4.25 µm |
| sensor dimensions (h,w) | 25.34 x 14.25 mm (diagonal 29 mm) | 21.49 x 12.61 mm (diagonal 24.9 mm) |
Pixel size of your images
The effective pixel size in acquired image depends on objective magnification you choose:
Pixel_size(image) = Pixel_size(sensor)/Magnification
| Magnification | mesoSPIM V6 pixel size, µm | Benchtop pixel size, µm |
| 1x | 5.5 | 4.25 |
| 2x | 2.75 | 2.125 |
| 5x | 1.1 | 0.85 |
| 7.5x | 0.73 | 0.567 |
| 10x | 0.55 | 0.425 |
| 20x | 0.275 | 0.212 |
Of course, the effective pixel size values are available in the meta-information files that accompany each dataset you acquire.
Field of view
Field of view can be calculated from the sensor size and objective magnifications by the formula:
FOV(h,w) = Sensor(h,w)/Magnification
Software (both systems)
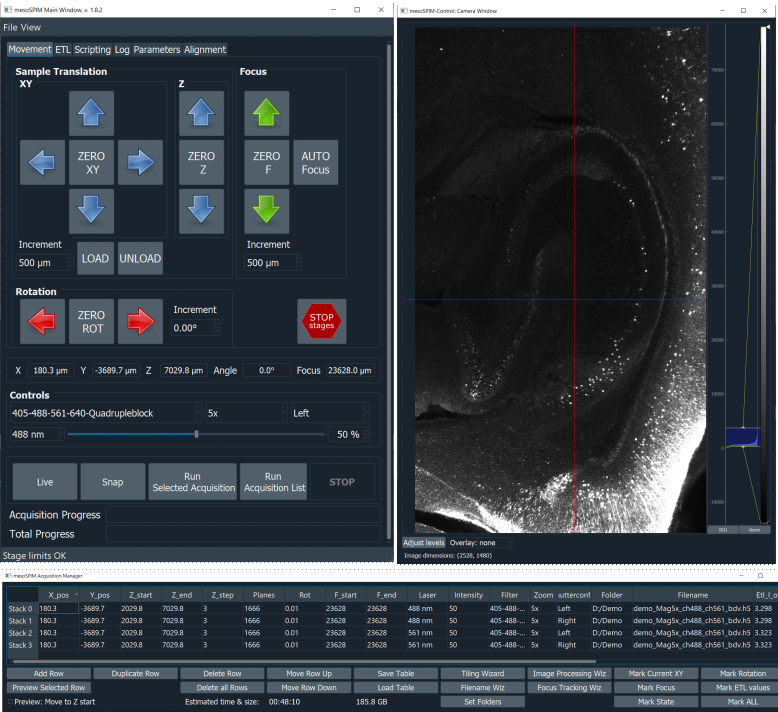
The microscope software, mesoSPIM-control, is developed by us. It is open-source and written in Python. It allows users to specify sequences of z-stacks using a table-based acquisition manager. The software can also be used to acquire large-scale tiling acquisitions.
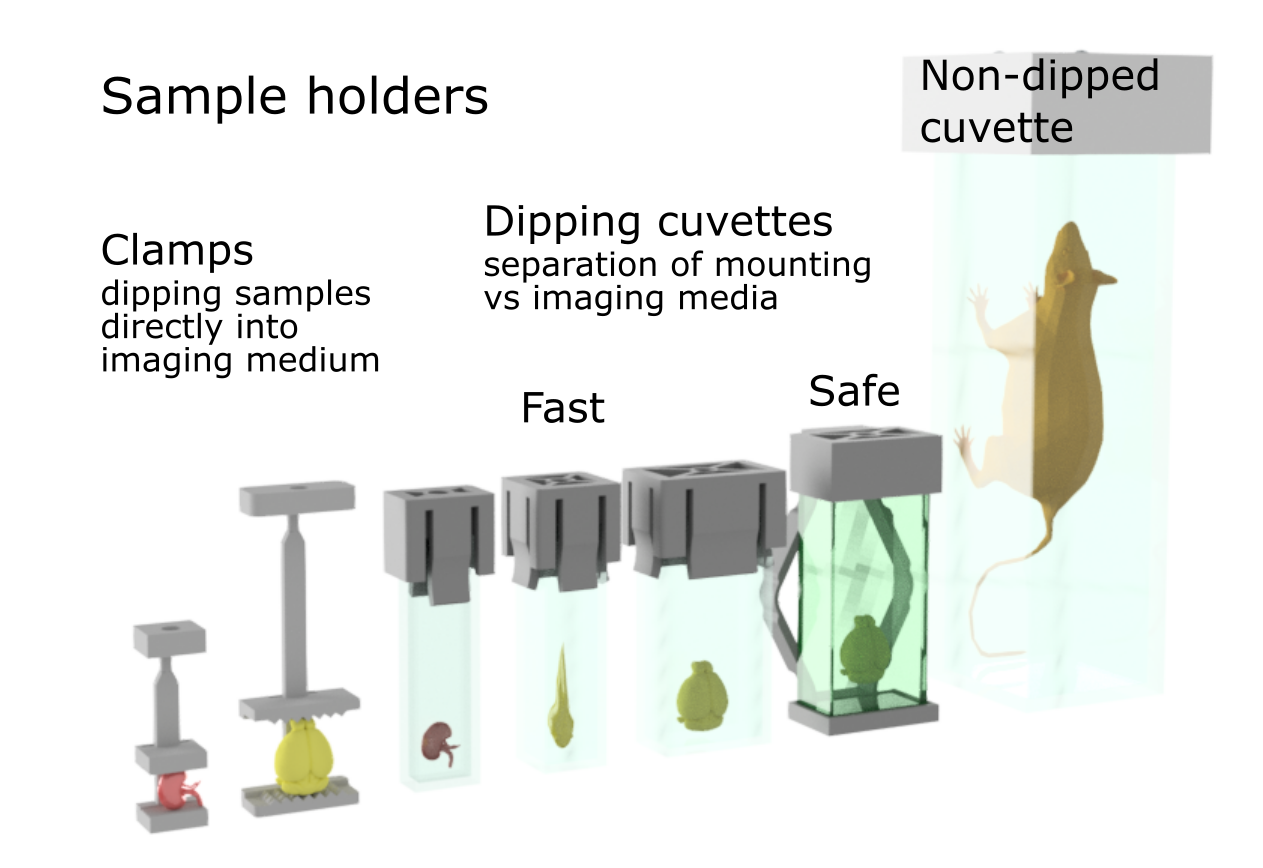
Depending on your sample, we have a variety of holders and imaging cuvettes.
Examples of samples imaged with Benchtop mesoSPIM
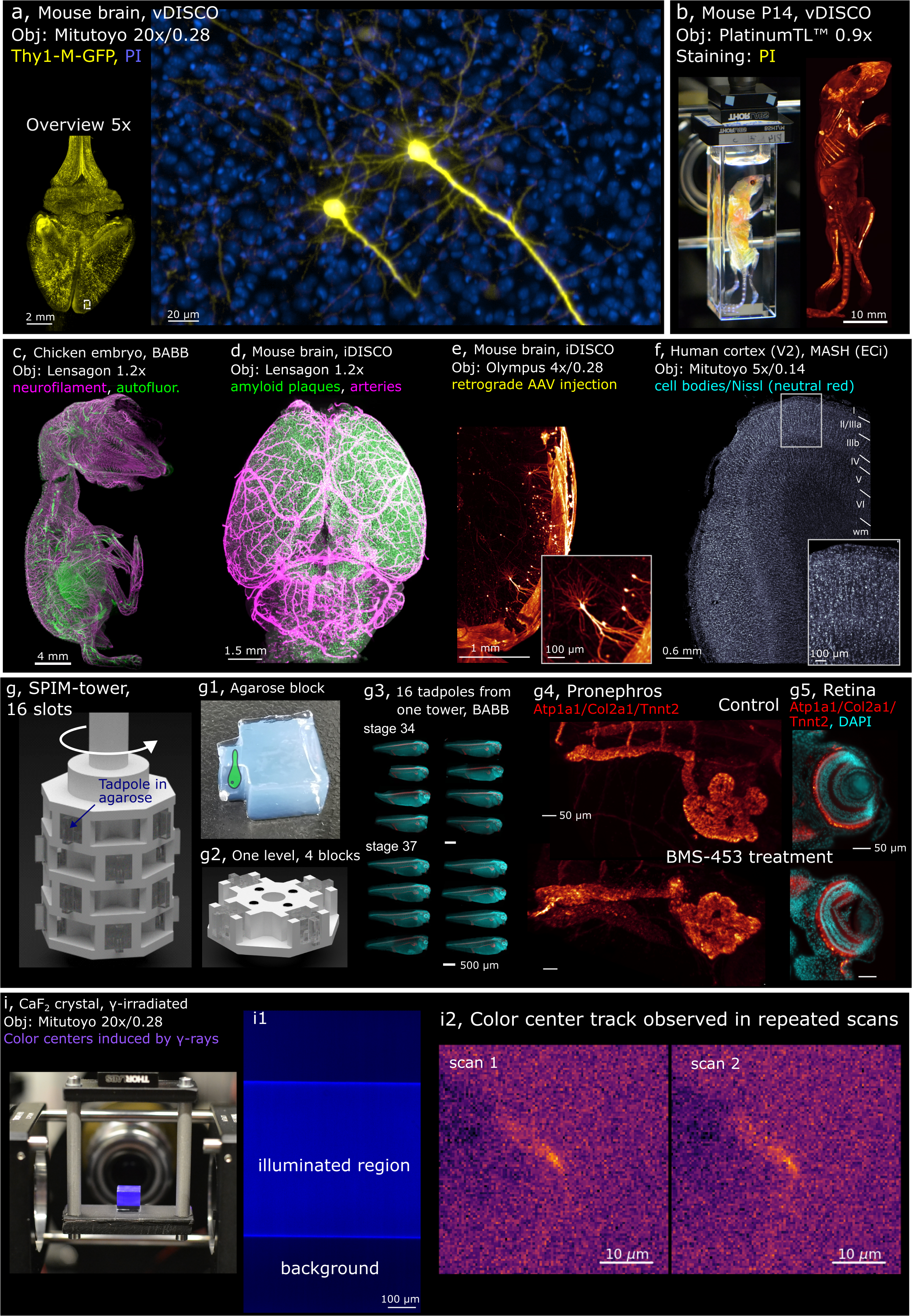
Differences between Benchtop and V6 mesoSPIMs
1. The way detection objectives can be exchanged
mesoSPIM V6: motorized objective turret ("revolver")
Benchtop: manual exchange (can be done without removing the sample).
2. Camera pixel size and total image size:
mesoSPIM V6: 5.5 µm pixel and 12 MP image.
Benchtop: 4.25 µm pixel and 15 MP image.
Other properties, such as lateral and axial resolution, maximum and minimum sample size, travel range of sample stages are nearly identical.
Custom accessories and unusual samples
For initial tests we provide standard accessories such as cuvettes, sample holders and immersion solutions. If you have unusual sample that requires customized accessories or supplies, talk us. We can suggest where to order custom accessories (such as ultra-long sample cuvettes), or even design and make them for you (such as specialized sample holders that are 3D printed form solvent-resistant polymers).
Literature and Links
Further information (internal UZH use only)
Follow this link for further background information, documents and links.
Responsible Persons
If you have questions about the device please contact the responsible person.
Acknowledgements
Make sure to acknowledge the Center for Microscopy in your publication
How to acknowledge contributions of the Center for Microscopy
The major upgrade of existing mesoSPIM to V6 "Revolver" in 2023 and the introduction of Benchtop mesoSPIM in 2024 was made possible with the generous support of URPP AdaBD Program.